
The Phage Receptor Binding Protein Database
A central hub for Tailspike and Tail Fibers
A web-based, searchable database for exploring phage receptor binding proteins (RBPs), including tailspikes and tail fibers. Built with Django and backed by MySQL, the platform enables filtering by taxonomy, domain architecture, and class. All proteins have experimental or AlphaFold/ESMFold predicted structures. Includes interactive visualization, export tools, and integrated domain annotations for in-depth analysis.
Database Features
Comprehensive tools for phage receptor binding protein research
Advanced Search
Filter by taxonomy, domain architecture, protein class, and evidence type to find exactly what you need.
3D Visualization
Interactive exploration of experimental and AlphaFold3-predicted protein structures with detailed annotations.
Data Export
Export search results in multiple formats for further analysis in your preferred tools and workflows.
Development Roadmap
Current initiatives and upcoming features
Phage Integration
- Integrate Phold into database
- Collect all phage-related proteins (GenBank + UniProt)
- Run sequence-based detection tools and import results
Prophage Pipeline
- Implement prophage pipeline
- Annotate prophage proteins and domains
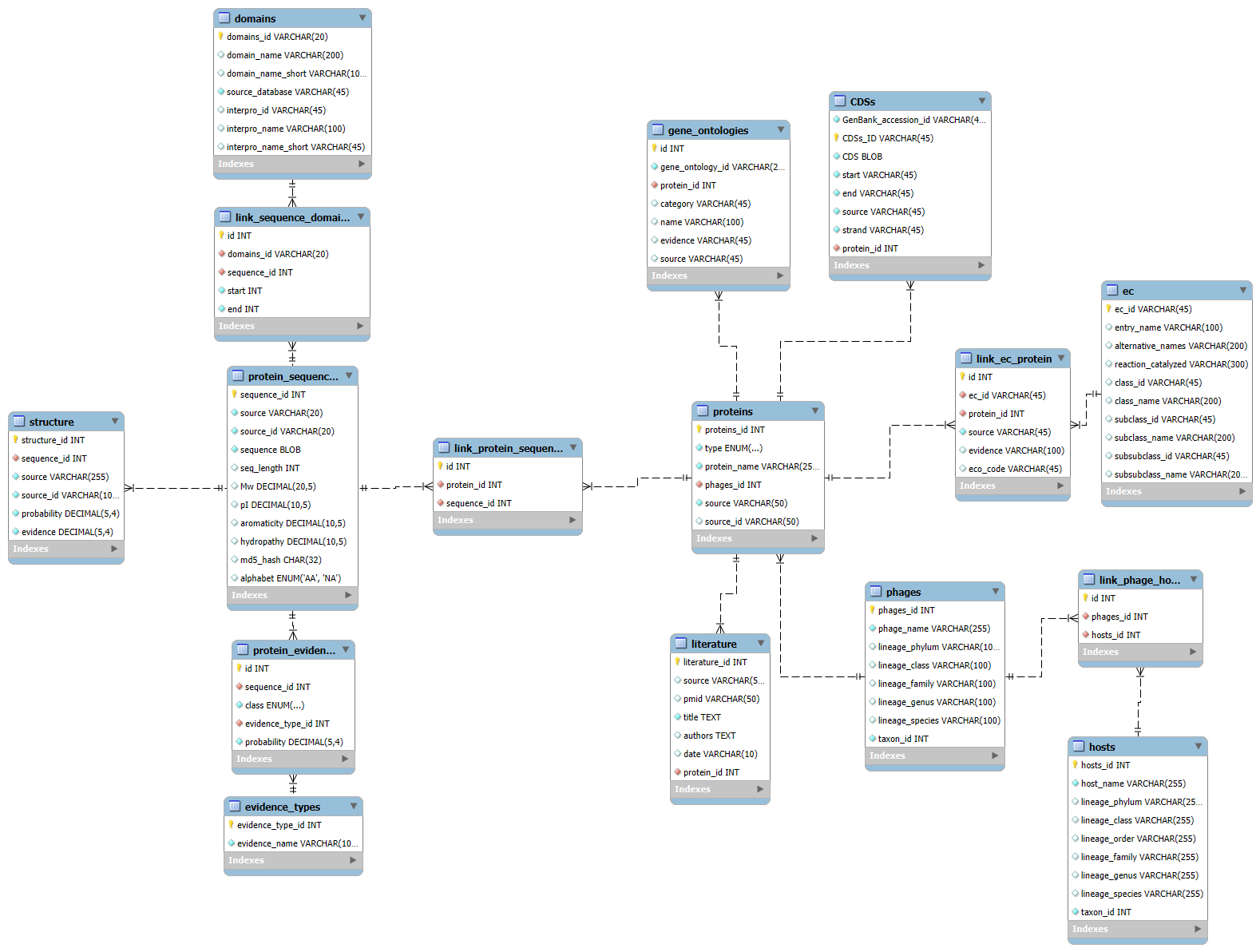
Relational Schema
Connected entities linking sequence and protein features.
The core PhaRP database centers arround protein metadata and sequence evidence. Proteins connect to phages, literature, enzymatic activity, while sequence data is unique linking to a unique stucture and predicted domains and evidence types.
- Non-reduntant with unique sequences and structures..
- Efficient filtering for protein properties and metadata.
- Composable annotations covering domains, ontology terms, and catalytic activity.
Evidence Integration
Comprehensive annotation from multiple bioinformatics tools
| Tool | Status | Tool | Status |
|---|---|---|---|
| UniProt annotation | ✔ Included | Genbank Annotation | ✔ Included |
| Phold | ✔ Included | DepoScope | ✔ Included |
| DePP | ✔ Included | PhageDPO | ✔ Included |
| RBPdetect2 | ✔ Included | RBPdetect | ✔ Included |
Data Collection Workflow
Automated pipeline for data integration and annotation

UniProt Pipeline
Fetch all phage taxons via NCBI using phage[Name Tokens], then retrieve UniProt entries with:
organism_id:{'{'}taxon_id{'}'} in FASTA format.
GenBank Pipeline
Filter GenBank UIDs with:
-db nucleotide -query "gbdiv_PHG[PROP] AND 1417:800000[SLEN]" | efetch -format uid
and download records for insertion.
Integration Steps
Insert proteins into database, merge evidence, fetch phage-host relationships, retrieve domain annotations, and populate literature metadata.
Run Locally
git clone https://github.com/victornemeth/PhaRBP.git
cd PhaRBP
docker compose up -d --build
# Access at http://localhost:8004Note: The official site is deployed via NPM; this repo is for development.